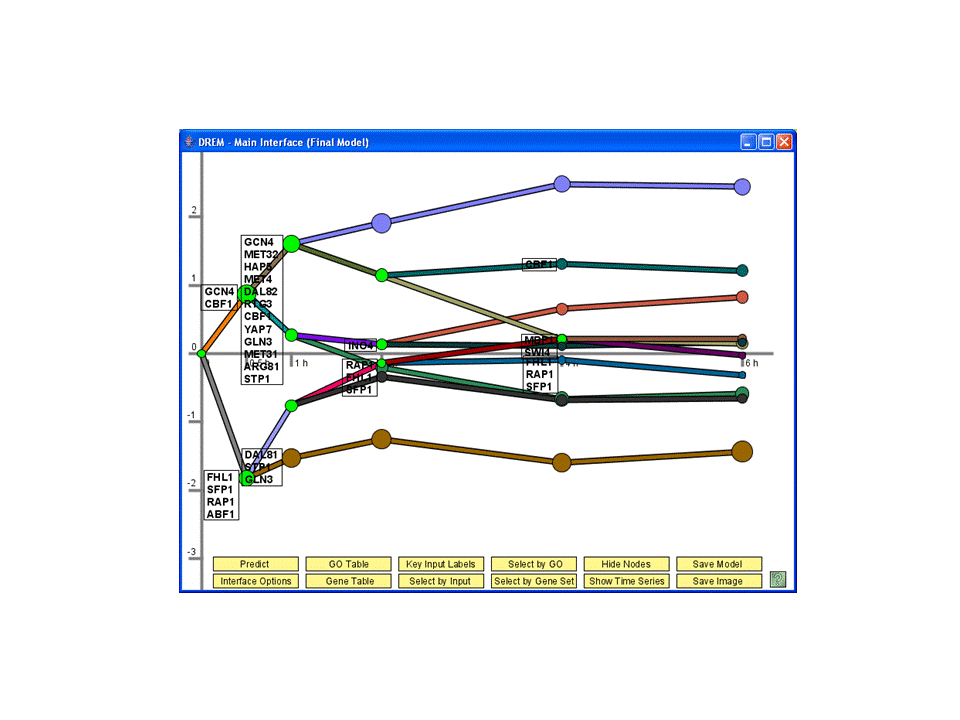
Regulatory Networks
Application of the Dynamic Regulatory Events Miner (DREM) to study amino acid starvation response in yeast.
Research
Our group develops computational methods for understanding the dynamics, interactions and conservation of complex biological systems. As new high-throughput biological data sources become available, they hold the promise of revolutionizing molecular biology by providing a large-scale view of cellular activity. However, each type of data is noisy, contains many missing values and only measures a single aspect of cellular activity. Our computational focus is on methods for large scale data integration. We primarily rely on machine learning and statistical methods. Most of our work is carried out in close collaboration with experimentalists. Many of the computational tools we develop are available and widely used.
Latest Publications
-
J. Hislop, Q. Song, K. Keshavarz., A. Alavi,. . ., Z. Bar-Joseph and M.R. Ebrahimkhani.
Modeling post-implantation human development to yolk sac blood emergence.
Nature , online ahead of print, 2023
-
M. Bailey, S. Moayedpour, . Li, A. Corrochano-Navarro, A. Kotter, L. Kogler-Anele, S. Riahi, C. Grebner, G. Hessler, H. Matter, M. Bianciotto, P. Mas, Z. Bar-Joseph*, and S. Jager*.
Deep Batch Active Learning for Drug Discovery.
eLife , 2023
-
S. Li, S. Moayedpour, R. Li, M. Bailey,. . ., Z. Bar-Joseph* and S. Jager*.
CodonBERT: Large Language Models for mRNA design and optimization.
Nuerips Workshop on Generative AI and Biology (GenBio@NeurIPS2023). , 2023
News
-
03/2021
Working with Yale and Pitt on the PF connectome We are leading the computational analysis and modeling efforts for a new project focused on Pulmonary Fibrosis (PF). Together with Yale and Pitt we are working on developing computational methods to model disease progression at the single cell level and match drugs to different disease stages to improve personalized treatment of PF. See the news story for more details.
-
10/2020
Chieh's thesis awarded SCS Honorable Mention Chieh's Lin PhD thesis, 'Probabilistic Single Cell Lineage Tracing', is one of 5 from the School of Computer Science at CMU selected for a PhD thesis award. Chieh is the second student from our group to receive this honor. Last year, Sabrina Rashid's thesis was also selected for a honorable mention (see news item below from 11/19). Given the large number of PhD students at SCS and the amazing work they all do, this is a great honor! Congratulations to Chieh.
-
06/2020
c3.ai award to develop AI methods for predicting cocktail treatments for SARS-COV-2. Our proposed project was one of a handful of proposal selected for funding by the C3.ai Digital Transformation Institute. Our project focuses on using machine learning methods to model the dynamic signaling and regulatory networks that are activated lung cells following infection with SARS-COV-2. Once such networks are reconstructed they will be analyzed to identify top proteins that can lead to reduction in viral loads. Next we will identify potential drugs to target combinations of these key proteins and test these on our recently constructed iPSC lung cell lines. This is joint work with our long time collaborator Darrell Kotton from Boston University and with Profs. Regina Barzilai and Tommi Jaakkola at MIT. See the c3.ai press release, the CMU story and a story in Pittsburgh Post-Gazette.